Recent News
Welcome to my website! I am committed to create a better world with precision cancer risk prediction and personalized medicine.
My research interests fall into four categories:
(1) Identifying genetic variants associated with cancer risk, with an emphasis on pancreatic cancer, using multi-omics approaches across diverse populations;
(2) Developing clinically applicable cancer risk assessment models;
(3) Developing drug response prediction models to guide personalized treatment;
(4) Uncovering key drivers of tumorigenesis, including somatic mutations and other contributing mechanisms.
Key words:
Computational Oncology (especially pancreatic cancer), Genome- and Transcriptome-Wide Association Study (GWAS/TWAS), Cancer Drivers, AI-powered Risk Assessment and Drug Response Prediction,Polygenic Risk Scores (PRS), Multi-Omics.
Employment
- Tenure-Track Assistant Professor (preparing) (2026.03 - present)
- Research Fellow (Full-time employee), National Cancer Institute, National Institutes of Health, US (2022 - 2026)
- Postdoctoral Fellow, National Cancer Institute, National Institutes of Health, US (2017 - 2022)
- Research Fellow (Full-time employee), University of Chinese Academy of Sciences,CN (2014 - 2017)
Education
- Ph.D. in Bioinfomatics, University of Chinese Academy of Sciences, CN (2009 - 2014)
- B. Sc. in Biotechology, South-Central University for Nationalities, CN (2005 - 2009)
Service
Journal Editor
- Editorial Board Member in BMC Medicine (2024 - present)
- Youth Editor in The Innovation (2022 - 2024)
- Associate Editor in Frontiers in Oncology (2021 - 2023)
Journal Reviewer (select):
- Gastroenterology, Nature Communications, The Innovation, Journal of Hematology & Oncology, Med, Genome Biology, eLife, BMC Medicine, Genomics Proteomics & Bioinformatics, Molecular Oncology, Cancer Letters.
All peer-review and editor records have been or are being certified by Web of Science.
Conference/Award Reviewer:
- American Society of Human Genetics (ASHG) annual meeting abstracts (2024)
- NIH DCEG Fellows Award for Research Excellence competition (DFARE) (2019)
Research
-
(I) Genome-Wide Association Studies (GWAS) to Identify Novel Susceptibility Variants for Pancreatic Cancer
A genome-wide association study (GWAS) is a methodology employed to link specific genetic variations with certain diseases. By identifying these variations, we gain insights into how variations/genes influence disease development, paving the way for enhanced prevention and treatment strategies. Focusing on pancreatic cancer susceptibility, my role encompasses leading the analyses within the Pancreatic Cancer Cohort Consortium (PanScan), a project under the auspices of the NCI-sponsored Cohort Consortium. Additionally, I am collaborating with the Pancreatic Cancer Case Control Consortium (PanC4) and the PANcreatic Disease ReseArch (PANDoRA) Consortium, aiming to bolster the detection power for identifying risk factors.
Related publications:
(1) Klein A, et al.Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nature Communications, 2018.
(2) Lin Y, et al.Genome-wide association meta-analysis identifies GP2 gene risk variants for pancreatic cancer. Nature Communications, 2020.
-
(II)Transcriptome-Wide Association Studies (TWAS) to Identify Novel Susceptibility Genes for Pancreatic Cancer
Most susceptibility variations discovered through GWAS reside in non-coding regions of the genome and likely function through variation-specific regulation of gene expression. A transcriptome-wide association study (TWAS) builds on this premise by imputing genetically predicted gene expression levels into GWAS datasets to discover genes whose cis-regulated expression is associated with complex traits.
Related publication:
(1) Zhong J, et al.A Transcriptome-Wide Association Study Identifies Novel Candidate Susceptibility Genes for Pancreatic Cancer. JNCI: Journal of the National Cancer Institute, 2020. PDF, Supp_PDF
-
[III]Cancer Risk Prediction
Related publication:
(1) Zhang, et al.Assessment of Polygenic Architecture and Risk Prediction based on Common Variants Across Fourteen Cancers. Nature Communications, 2020.
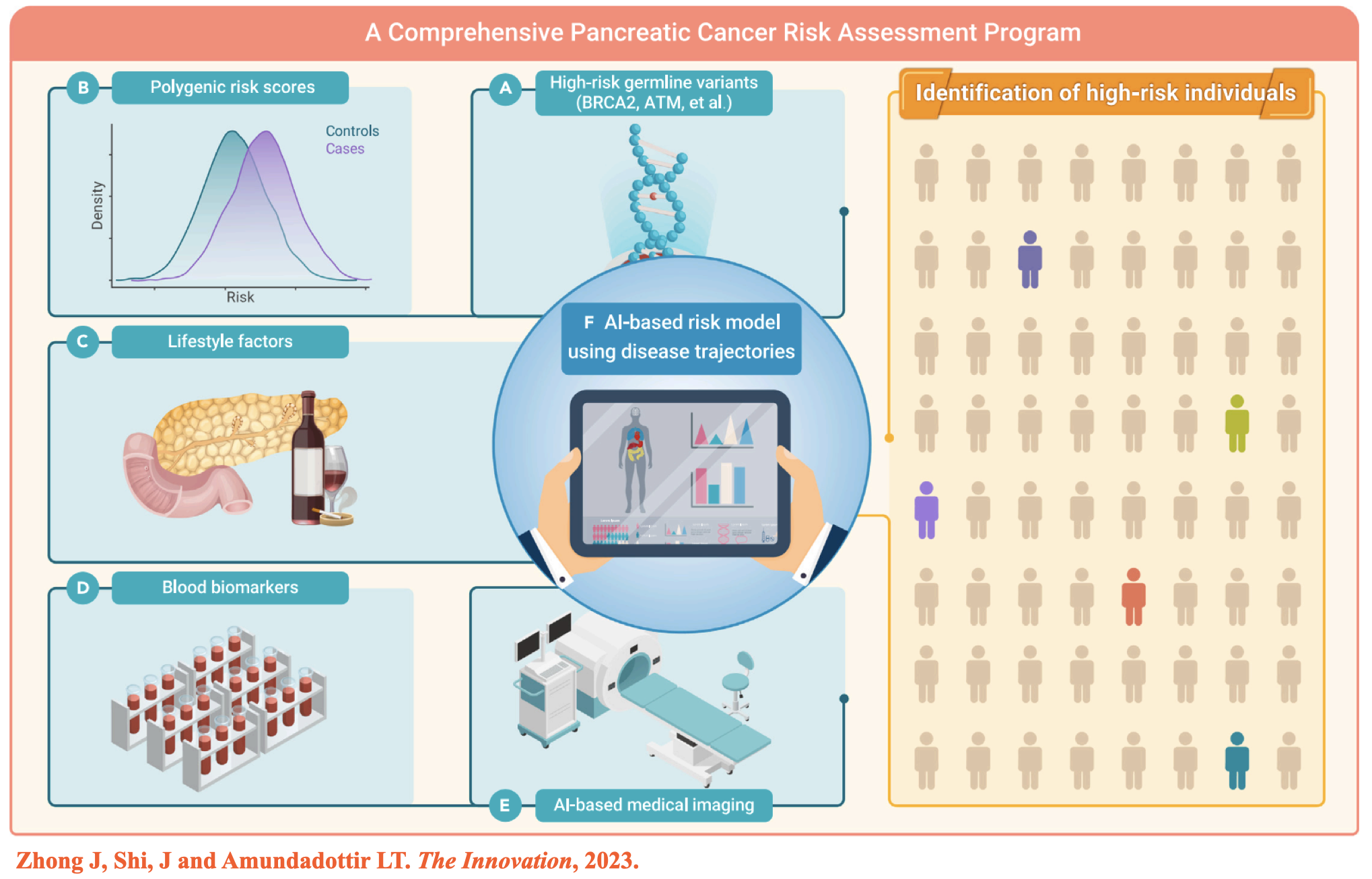
(2) Zhong J, et al. Artificial Intelligence and improved early detection for pancreatic cancer. The Innovation, 2023. -
[IV] Somatic driver mutations/epigenetic drivers for tumorgenousis
Related publications:
(1) Zhong J et al. Large-scale multi-omic analysis identifies noncoding somatic driver mutations and nominates ZFP36L2 as a driver gene for pancreatic ductal adenocarcinoma. Gut, 2025.
(2) Zhong J, Amundadottir LT. Uncovering dark matter in cancer by identifying epigenetic drivers. Trends in Genetics, 2024. -
[V] Fine Mapping for Pancreatic Cancer Risk
Fine-mapping is the process by which a trait-associated region from a GWAS is analysed to identify the particular genetic variants that are likely to causally influence the examined trait.
Related publications:
(1) Jermusyk A#, Zhong J#, et al.A 584 bp deletion in CTRB2 inhibits chymotrypsin B2 activity and secretion and confers risk of pancreatic cancer.AJHG, 2021. PDF, Supp
-
Other whole genome and methyome sequencing projects
Related publications:
Huang H, et al. Clinical Infectious Diseases, 2018; Yang T#, Zhong J#, et al. Frontiers in Microbiology, 2018; Zhu L#, Zhong J#,et al. Nucleic Acids Research, 2016; Wang F#, Zhong J#, et al. BMC Genomics, 2014.
Selected Publications
# co-first author, § co-corresponding author:
- Zhong J§, et al, Amundadottir LT§. Large-scale multi-omic analysis identifies noncoding somatic driver mutations and nominates ZFP36L2 as a driver gene for pancreatic ductal adenocarcinoma. Gut, 2025.
- Zhong J§, Shi J§, Amundadottir LT§. Artificial Intelligence and improved early detection for pancreatic cancer. The Innovation, 2023.
- Jermusyk A#, Zhong J#,et al. A 584 bp deletion in CTRB2 inhibits chymotrypsin B2 activity and secretion and confers risk of pancreatic cancer. The American Journal of Human Genetics, 2021. PDF, Supp
- Zhong J, et al. A Transcriptome-Wide Association Study Identifies Novel Candidate Susceptibility Genes for Pancreatic Cancer. JNCI: Journal of the National Cancer Institute, 2020. [PDF], [Sup_PDF] (Highlighted with an editorial in JNCI)
- Zhu L#, Zhong J#, et al. Precision methylome characterization of Mycobacterium tuberculosis complex using PacBio single-molecule real-time technology. Nucleic Acids Research, 2016.
Other Publications
For journals (# for co-first author, § for corresponding author):
- Connelly KE, […] Zhong J, et al. Allelic effects on KLHL17 expression likely mediated by JunB/D underlie a PDAC GWAS signal at chr1p36.33. Nature Communications. 2025
-
Zhong J§, Amundadottir LT§. Uncovering dark matter in cancer by identifying epigenetic drivers. Trends in Genetics, 2024.
-
Ni Z, […] Zhong J, et al. Genome-wide analysis to assess if heavy alcohol consumption modifies the association between SNPs and pancreatic cancer risk. Cancer Epidemiology, Biomarkers & Prevention, 2024.
- King S, […] Zhong J, et al. Genetic susceptibility to nonalcoholic fatty liver disease and risk for pancreatic cancer: Mendelian randomization. Cancer Epidemiology, Biomarkers & Prevention, 2023.
-
Lindström S, […] Pancreatic Cancer Cohort Consortium (Panscan)(including Zhong J), et al. Genome-Wide Analyses Characterize Shared Heritability Among Cancers and Identify Novel Cancer Susceptibility Regions. JNCI: Journal of the National Cancer Institute, 2023.
- Hoskins JW, […] Zhong J, et al. Inferred expression regulator activities suggest genes mediating cardiometabolic genetic signals. PLoS Computational Biology, 2021.
-
Julian-Serrano S, […] Zhong J, et al. Hepcidin-regulating iron-metabolism genes and pancreatic ductal adenocarcinoma, a pathway analysis of genome-wide association studies. The American Journal of Clinical Nutrition, 2021.
-
Zhang YD, […] Pancreatic Cancer Cohort Consortium (PanScan)(including Zhong J) et al. Assessment of Polygenic Architecture and Risk Prediction based on Common Variants Across Fourteen Cancers. Nature Communications, 2020.
- Klein AP, […] Zhong J, et al. Genome-wide meta-analysis identifies five new susceptibility loci for cancer. Nature Communications, 2018.
- Huang H, […] Zhong J, et al. Cross-sectional Whole-genome Sequencing and Epidemiological Study of Multidrug-resistant Mycobacterium tuberculosis in China. Clinical Infectious Diseases, 2018.
-
Yang T#, Zhong J#, et al. Pan-genomic study of Mycobacterium tuberculosis reflecting the primary/secondary genes, generality/individuality, and the interconversion through copy number variations. Frontiers in Microbiology, 2018.
- Zhang L#, Zhong J#, et al. Complete genome sequence of the drought resistance-promoting endophyte Klebsiella sp. LTGPAF-6F. Journal of Biotechnology, 2017.
- The IC4R Project Consortium (including Zhong J). Information Commons for Rice. Nucleic Acids Research, 2016.
- Wang FQ#, Zhong J#, et al. Genome sequencing of high-penicillin producing industrial strain of Penicillium chrysogenum. BMC Genomics, 2014.
- Ling Y, […] Zhong J, et al. VCGDB: a dynamic genome database of the Chinese population. BMC Genomics, 2014.
- Wu J, […] Zhong J, et al. Systematic analysis of intron size and abundance parameters in diverse lineages. Sci China Life Sci, 2013.
For conferences:
- O’Brien A, […] Zhong J, et al. Uncovering the functional variants and target genes of the 7q32 pancreatic cancer risk locus. Proceedings of the American Association for Cancer Research Virtual Special Conference on Pancreatic Cancer (AACR). Cancer Research, 2022.
- Jermusyk A, Zhong J, et al. An exception to the rule: A coding functional variant at a pancreatic cancer GWAS locus. Proceedings of the American Association for Cancer Research Virtual Special Conference on Pancreatic Cancer (AACR). Cancer Research, 2020.
- Zhong J, et al. Large-scale transcriptome-wide association study identifies novel candidate susceptibility genes for pancreatic cancer. Proceedings of the American Association for Cancer Research Annual Meeting (AACR). Cancer Research, 2019.
Presentations
Invited speaker
-
Identifying noncoding somatic driver mutations in pancreatic cancer. Presented at NCI DCEG Early Career Scientists seminar, virtually, March 2023.
-
Pancreatic Cancer Cohort Consortium (PanScan IV) Update. Presented at Pancreatic Cancer Case-Control Consortium (PanC4) virtual annual meeting, virtually, December 2022.
-
Identifying non-coding somatic driver mutations in regulatory elements for pancreatic cancer. Presented (Platform) at American Society of Human Genetics (ASHG) annual meeting, Los Angeles, October 2022.
-
Pancreatic Cancer Cohort Consortium (PanScan) IV GWAS update. Presented in the FinnGen Scientific Committee/Analysis group meeting, virtually, January 2022
-
PanScan update and preliminary results for PANDoRA and Asian GWAS scans. Presented in the PanScan cohort consortium annual meeting, virtually, October 2021.
-
Uncovering the dark matter of pancreatic cancer: noncoding somatic driver mutations in regulatory elements. Presented in the CRL (Consolidated Research Laboratory) seminar, Division of Cancer Epidemiology and Genetics (DCEG, NCI), virtually. September 2020.
-
Large-scale Transcriptome-Wide Association Study (TWAS) Identifies Novel Candidate Susceptibility Genes for Pancreatic Cancer. Presented in the 11th Annual Division of Cancer Epidemiology and Genetics (DCEG, NCI) Fellows’ Training Symposium, Bethesda, MD. April 2019.
-
A Transcriptome-Wide Association Study (TWAS) Identifies Novel Candidate Susceptibility Genes for Pancreatic Cancer. Presented at the LTG (Laboratory of Translational Genomics) seminar, Division of Cancer Epidemiology and Genetics (DCEG, NCI), Gaithersburg, MD. February 2019.
-
Genome sequencing of high-penicillin producing industrial strain of Penicillium chrysogenum. Invited Speaker at the twelfth Asia Pacific Bioinformatics Conference, Shanghai, China. January 2014. (Best Paper Award in the conference)
Selected Posters:
-
Zhong J, O’Brien A, Patel M, Eiser D, Mobaraki M, Collins I, Wang L, Guo K, TruongVo T, Jermusyk A, O’Neill M, D Dill CD, Wells AD, Leonard ME, Pippin JA, Grant SFA, Zhang T, Andresson T, Connelly KE, Shi J, Arda HE, Hoskins JW, Amundadottir LT. Large-scale multi-omic analysis identifies noncoding somatic driver mutations and nominates ZFP36L2 as a driver gene for pancreatic ductal adenocarcinoma. Presented at the American Association for Cancer Research Annual Meeting, Chicago, IL. April 2025.
-
Zhong J, Eiser D, Brien A, Connelly K, Hoskins J, Jermusyk A, Collins I, Shen T, Zhao Y, Li Wang, Guo K, TruongVo T, Wildenthal J, McKinnon K, Wells A, Grant S, Arda E, Shi J, Amundadottir L. Uncovering the dark matter of pancreatic cancer: identifying driver mutations in non-coding gene regulatory elements. Presented at American Society of Human Genetics virtual meeting (ASHG), October 2020. [Reviewers’ Choice award, top 10%]
-
Zhong J, Jermusyk A, Wu L, Hoskins J, Collins I, Zhang M, Lei S, Chung C, Zhang T, Xiao W, Stolzenberg-Solomon R, Klein A, Wolpin B, Shu X, Chanock S, Olson S, Chatterjee N, Smith J, Jianxin Shi, Kraft P, Petersen G, Zheng W, Amundadottir L. Large-scale transcriptome-wide association study (TWAS) identifies novel candidate susceptibility genes for pancreatic cancer. Presented at American Association for Cancer Research Annual Meeting (AACR), Atlanta, GA. March 2019.
-
Zhong J, Wu L, Jermusyk A, Hoskins J, Collins I, Zhang M, Lei S, Chung C, Zhang T, Xiao W, PanScan, PanC4, Stolzenberg-Solomon R, Klein A, Wolpin B, Brown K, Shu X, Chanock S, Olson S, Chatterjee N, Petersen G, Smith J, Shi J, Kraft P, Zheng W, Amundadottir L. A Transcriptome-wide association study for pancreatic cancer. Presented at American Society of Human Genetics annual meeting (ASHG), San Diego, CA. October 2018.
Awards
- DCEG Intramural Research Award ($50,000), National Cancer Institute (2021)
- Fellows Award for Research Excellence, National Institutes of Health (2021)
- Reviewers’ Choice Poster Award (Top 10%), American Society of Human Genetics (ASHG) annual meeting (2020)
- 2021 NCI Directors Innovation Award (Participant) (2020)
- Fellows Award for Research Excellence, National Institutes of Health (2019)
- Summer Research Mentor Award, National Institutes of Health (2019)
- Visiting Fellowship Award, National Cancer Institute, National Institutes of Health (2017 - 2022)
- Best Paper Award, Asia Pacific Bioinformatics Conference (2014)
- Merit Student (Top 15%), University of Chinese Academy of Sciences (2011)
- Merit Student (Top 15%), University of Chinese Academy of Sciences (2010)
Professional Membership/Role
- Member in American College of Medical Genetics and Genomics (ACMG) (2024)
- Gold Member in American Association for the Advancement of Science (AAAS) (2023 - 2024)
-
Member in American Society of Human Genetics (ASHG)(2022-2024)
- Associate Member in American Association for Cancer Research (AACR)(2018-2020;2024-2025)
- Member in Tumor Microenvironment Group of the AACR (2018-2019)
- Planning Committee Member in 10th Annual NCI DCEG Fellows’ Training Symposium (2017-2018)
- Member in American Society of Human Genetics (2017-2018)
Contact
- joshchung@foxmail.com
Posts
- 05/18/2025: updated
- 10/03/2024: updated
- 05/18/2024: updated
- 12/08/2023: updated
- 06/05/2023: updated
- 05/25/2021: my website is under construction and will be updated!
